Research
Human developmental disease
Faithful epigenetic regulation of the genome is critical for normal development. Disruption to these processes, for instance through genetic mutations in epigenetic regulators, can result in an altered distribution of chromatin modifications, either localized or genome-wide and human disease. We aim to understand how altered chromatin states influence gene expression patterns in clinically relevant cell types and identify endogenous targets amenable for (epigenetic) therapy.
Epigenetic regulator function
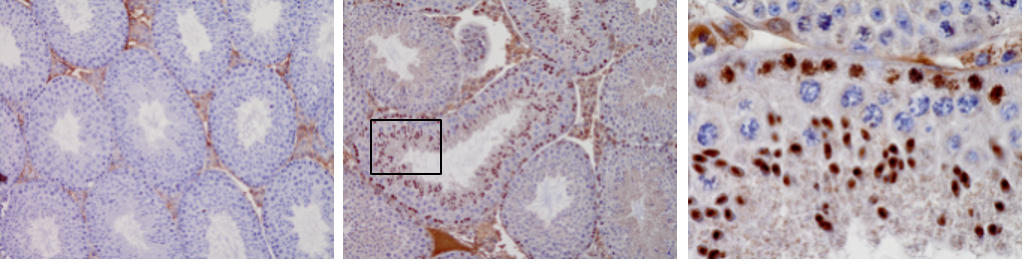
Our previous work identified a set of ~30 unique genes from an in vivo chemical mutagenesis screen designed and performed in Emma Whitelaw’s lab. They are known as Modifiers of murine metastable epialleles Dominant (MommeD), and contain members of the DNA methylation, histone modification and chromatin remodeling pathways. We use this exquisite resource to study epigenetically regulated processes including transposable element silencing and the regulation of complex genomic structures such as satellite repeats and large gene clusters. A current focus of the lab are neurodevelopment and immune cell development and function.
Epigenetic inheritance
In mammals, epigenetic marks are usually cleared on passage through the germline. This is to ensure the totipotency of the zygote and serves to erase altered epigenetic states that may have arisen in the parental generation (e.g., through environmental influences). However, an increasing number of reports that (altered) epigenetic state can be inherited to the next generation via the gametes and influence offspring phenotype, challenges this dogma. We aim to identify genomic loci (metastable epialleles) that are particularly sensitive to outside influences.